Comparative Genomics to Investigate the Resistome and Bacterial Heterogeneity in the Creekside Ecosystem in Navotas City: An Exploratory Research
DOI:
https://doi.org/10.65232/rcdf2g95Keywords:
Antibiotic Resistance Gene, Antimicrobial Resistance, Creekside Bacteria, Public Health, Shotgun Metagenomic SequenceAbstract
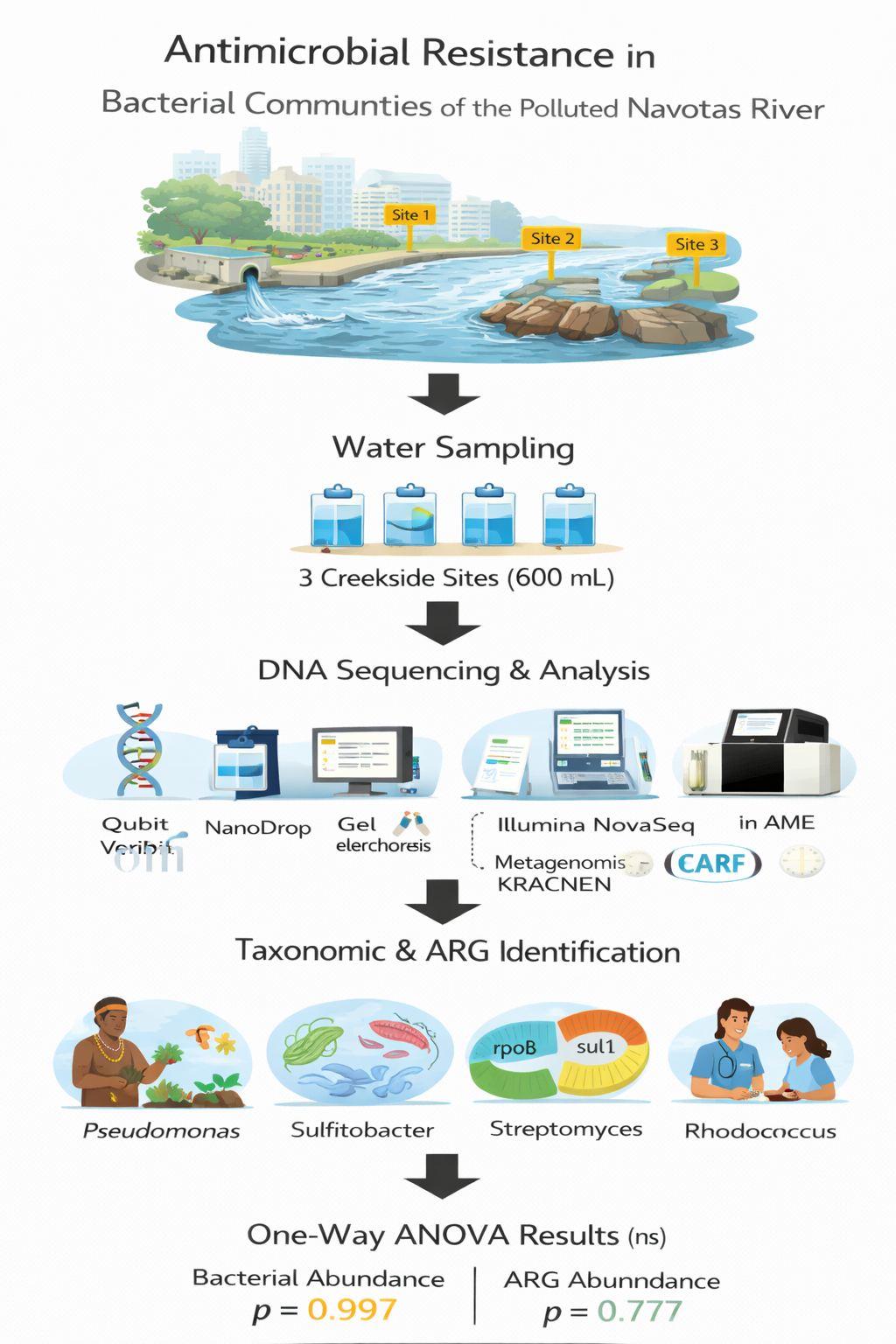
Rapid urbanization and untreated sewage discharge have degraded aquatic ecosystems, particularly the Navotas River. This study aimed to assess antimicrobial resistance (AMR) in bacterial communities to support rehabilitation efforts. A descriptive-correlational design was employed at three creekside sites with varying levels of human activity. Triplicate water samples (600 mL/site) were collected during the first and fourth weeks of January 2025. DNA was extracted and validated using Qubit, NanoDrop, and gel electrophoresis. Shotgun metagenomic sequencing was performed via Illumina NovaSeq (15 million reads/sample). Taxonomic classification used QIIME and KRAKEN, while antimicrobial resistance genes (ARGs) were identified through Q2-RGI with the comprehensive antimicrobial resistance database (CARD). One-way ANOVA assessed differences in bacterial and ARG abundance across sites. DNA quality met sequencing requirements, and physicochemical parameters, including pH and temperature were within normal limits. A diverse microbial community with known pathogenicity was found, including Pseudomonas, Sulfitobacter, Streptomyces, and Rhodococcus. The most abundant ARG was rpoB mutant in Bifidobacterium adolescentis (35.41%), conferring rifampicin resistance, followed by rpoB2 in Nocardia (20.94%). Other notable ARGs included sul1 (8.61%) and tlrC (4.90%), which are associated with sulfonamide and macrolide-lincosamide resistance. Beta-lactamases, including VIM, NDM, OXA variants, including aminoglycoside-resistance genes were identified. No significant ARG abundance differences were found across sites (p = 0.997; p = 0.777). The presence of antibiotic-resistant bacteria and ARGs, particularly rpoB mutants and beta-lactamases, signals a silent AMR crisis in Navotas River. These findings highlight the urgent need for policy intervention and continuous AMR surveillance in aquatic environments.
Downloads
References
Edullantes, A. M., Cagurin, G. B., & Walag, A. M. P. (2024). A community project on waste management awareness and livelihood training of residents nearby a polluted creek: An impact study. Environment and Ecology Research, 12(2), 172-180. https://doi.org/10.13189/eer.2024.120208
Khine, N. O., Lugsomya, K., Kaewgun, B., Honhanrob, L., Pairojrit, P., Jermprasert, S., & Prapasarakul, N. (2020). Multidrug resistance and virulence factors of Escherichia coli harboring plasmid-mediated colistin resistance: mcr-1 and mcr-3 genes in contracted pig farms in Thailand. Frontiers in Veterinary Science, 7, 582899. https://doi.org/10.3389/fvets.2020.582899
Liu, D., Zhou, H., Xu, T., Yang, Q., Mo, X., Shi, D., Ai, J., Zhang, J., Tao, Y., Wen, D., Tong, Y., Ren, L., Zhang, W., Xie, S., Chen, W., Xing, W., Zhao, J., Wu, Y., Meng, X., ... Wang, Y. (2021). Multicenter assessment of shotgun metagenomics for pathogen detection. EBioMedicine, 74, 103649. https://doi.org/10.1016/j.ebiom.2021.103649
Lunha, K., Leangapichart, T., Jiwakanon, J., Angkititrakul, S., Sunde, M., & Järhult, J. D. (2020). Antimicrobial resistance in fecal Escherichia coli from humans and pigs at farms at different levels of intensification. Antibiotics, 9(10), 662. https://doi.org/10.3390/antibiotics9100662
Numberger, D., Zoccarato, L., Woodhouse, J., Ganzert, L., Sauer, S., Márquez, J. R. G., Domisch, S., Grossart, H., & Greenwood, A. D. (2022). Urbanization promotes specific bacteria in freshwater microbiomes including potential pathogens. The Science of the Total Environment, 845, 157321. https://doi.org/10.1016/j.scitotenv.2022.157321
Sihombing, B., Bhatia, R., Srivastava, R., Aditama, T. Y., Laxminarayan, R., & Rijal, S. (2023). Response to antimicrobial resistance in South-East Asia region. The Lancet Regional Health - Southeast Asia, 18, 100306. https://doi.org/10.1016/j.lansea.2023.100306
Singh, S. (2020). Conventional infection prevention and control practices in post-antibiotic era: A perspective. Journal of Scientific Research, 64(01), 167–174. https://doi.org/10.37398/jsr.2020.640124
Additional Files
Published
Issue
Section
License
Copyright (c) 2025 Mariae Janselle Pamposa, Zamantha Bernadette Convento, Jhoanna Loraine Fabela, Khris Angelique Manalo, Duane Miguel Mariano, Eulce Jashen Regala, Hana Yesha Uthayasurian, Maria Ruth Pineda-Cortel, Sherill Tesalona (Author)

This work is licensed under a Creative Commons Attribution 4.0 International License.
Articles in the APCORE Online Journal (AOJ) are published under the Creative Commons Attribution 4.0 International License (CC BY 4.0). Authors retain copyright and grant the journal the right of first publication, allowing others to share and adapt the work with proper attribution to the original author and source.
https://creativecommons.org/licenses/by/4.0/




